Similar to how a bucket can be used to save a sinking boat, some cells contain unique technology that allows them to evacuate pollutants. Unfortunately, antibiotic treatments can be considered “toxins.”
A Cornell-led team has discovered the machinery that allows bacteria to withstand antibiotic exposure: a shuttling process that facilitates the pumping out of a variety of antibiotics and other physiological substrates from the cell by a complex of proteins.
With the mechanism now known, scientists are attempting to sabotage the process with chemical and mechanical means to allow antibiotics to function without hindrance.
Their findings were published in the journal Cell Reports Physical Science.
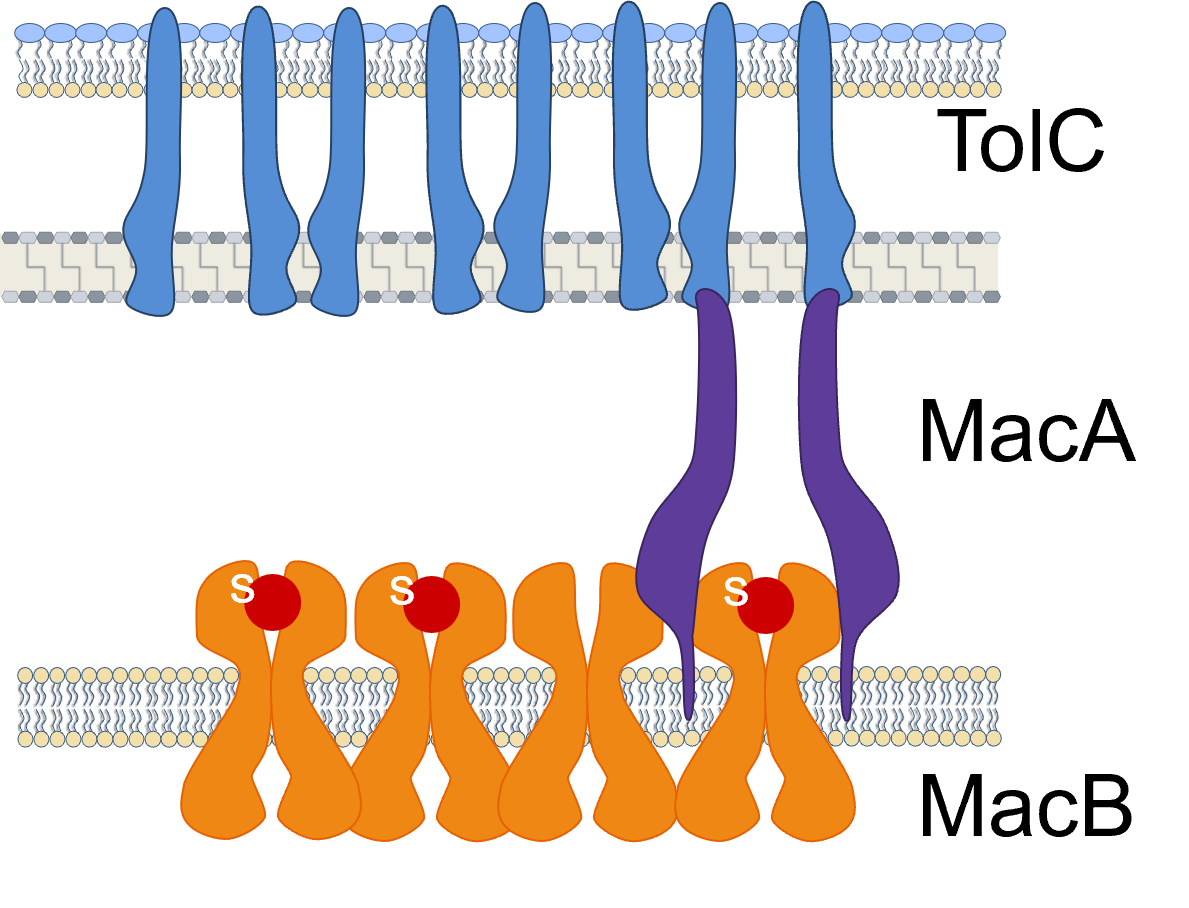
Antibiotic resistance varies among bacteria, with so-called gram-negative bacteria being especially robust because to their additional membrane for self-argument. A three-part protein complex called MacAB-TolC, which spans the cell’s inner and outer membranes as well as the periplasm that joins them, serves as its intricate plumbing system. Despite being anchored to the inner membrane, MacA is found in the periplasm, TolC is found on the outer membrane, and MacB is found on the inner membrane.
This “tripartite” protein complex, sometimes referred to as a multidrug efflux pump, creates a channel that drains out virulence factors, or compounds made by the bacterial cell itself, in addition to antibiotics.
The three proteins must come together in a particular stoichiometry, or chemical equilibrium, in order to pump out poisons. Six MacA proteins and two MacB proteins come together to form three TolC proteins. Although that ratio is well known, scientists have been curious about how molecules in the periplasm enter the channel that passes through the complex and what kinds of substrates they are pumped through once the structure is put together.
To gain a deeper understanding of the protein interactions, Chen’s team employed single-molecule imaging on E. coli bacterial cells. The stoichiometry was actually out of balance, with many more MacBs floating around (and even more TolC, which was known from earlier studies) than were required for the complex’s 2:6:3 shape, they discovered when they analyzed the protein concentration inside the cell. Additionally, scientists observed that the adaptor protein MacA might separate from the MacAB-TolC assembly.
You basically have these extra Bs that don’t have A partners to assemble. And of course, the cell does not do this for no reason,
We found out a good reason is that when you have this extra B, because it does not have A associated with it, it naturally has an opening for the substrate to go in. So once the substrate can bind to the extra B, some of the A’s that are initially associated with B can migrate over to assemble. And once it’s assembled, they can pump the substrate out.
Peng Chen
Chen’s group worked with a team at the University of California, San Francisco, led by former Cornell professor Christopher Hernandez, to see if the mechanism could be disrupted. Hernandez created a microfluidic device that can alter a bacterium’s resistance to toxins by applying mechanical stress.
Squeezing E. coli through the apparatus caused enough cell deformation, the researchers found, to upset the complex that had been put together and stop it from being resistant to drugs.
Now that he has established the link between cellular function and unbalanced protein stoichiometry, Chen is interested in seeing if this relationship holds true for other systems.
This imbalance of protein stoichiometry must exist for many types of protein complexes. But how does a cell utilize this imbalance? I don’t think people realize this sufficiently,
Now we have one example that shows this particular imbalance might be, functionally, very relevant. So anytime that we study protein complexes in the cell, we always want to measure the relative amount in the entire cell versus their relative amount in a particular complex. Do they actually match?
Peng Chen
Source: Cornell Chronicle
Journal Reference: Wenyao Zhang, Christine E. Harper, Junsung Lee, Bing Fu, Malissa Ramsukh, Christopher J. Hernandez, Peng Chen. Transporter excess and clustering facilitate adaptor protein shuttling for bacterial efflux. Cell Reports Physical Science, 2025; 102441 DOI: 10.1016/j.xcrp.2025.102441
Last Modified: