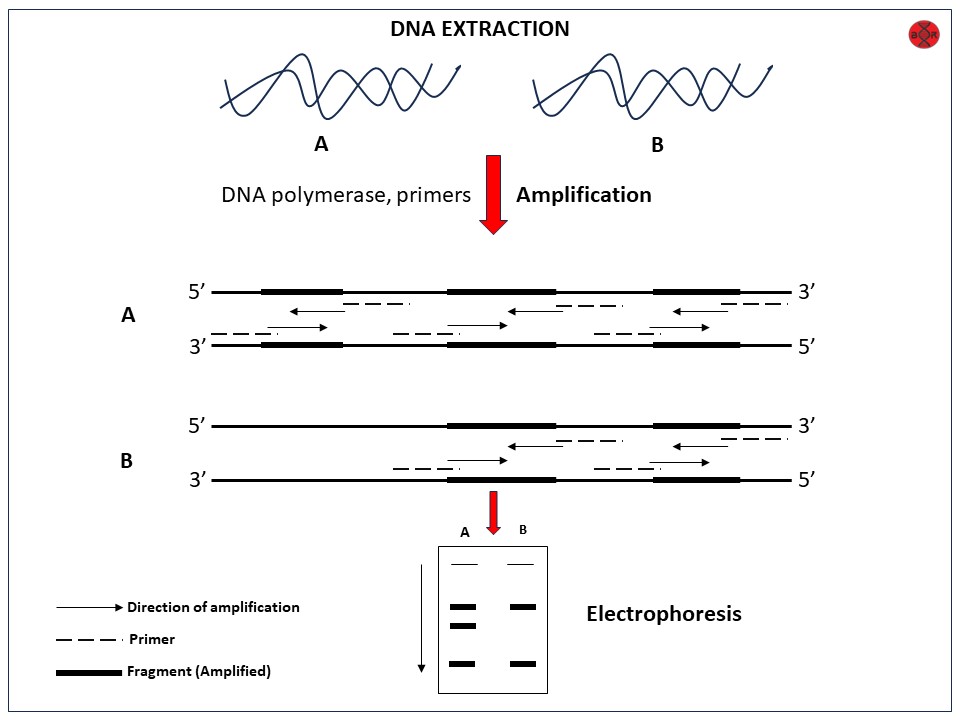
The basis of the randomly amplified polymorphic DNA (RAPD) technique is differential PCR amplification of genomic DNA. It uses short random oligonucleotide sequences (usually 10 bases long) to infer DNA polymorphisms caused by “rearrangements or deletions at or between oligonucleotide primer binding sites in the genome.”
The method can be used across species by utilizing universal primers because it necessitates no prior knowledge of the genome being studied. In the RAPD marker system, a PCR reaction is carried out using a single RAPD primer and just a small amount of template DNA (even less than 10 ng is adequate).
Primers often have a random sequence and are just 10 base pairs long (10 meters). The RAPD study is based on the PCR employing single, short (approximately 10 bases) randomly chosen primers that anneal as reversed repeats to the complementary sites in the genome.
PCR uses the primers as the starting and ending points to amplify the DNA between two opposing sites. In the presence of ethidium bromide, the amplification products are separated on agarose gels and examined under ultraviolet light. Polymorphism in the banding patterns allows for the differentiation of organisms based on the presence or absence of bands.
Although length differences in the amplified sequence between primer annealing sites can potentially cause these polymorphisms, variations in the primer annealing sites are thought to be the primary cause.
Each product is generated from a genome region with two short sections reversed in their orientation and complementary to the primer on opposing strands.
The characteristic of RAPD analysis is that it makes distinctions at several taxonomic levels, i.e., isolates and species, depending on the organism under examination and the primer employed.

Advantages:
The primary benefit of RAPDs is their rapidity and ease of assay. Since PCR is involved, very small amounts of template DNA—typically 5–50 ng per reaction—are needed. There is no need for sequence data for primer synthesis because random primers are commercially accessible.
Additionally, RAPDs are dispersed at random throughout the entire genome and have a very high genomic abundance. They have limits when used as mapping markers because they are dominating markers, however, these restrictions can be partially mitigated by choosing markers that are linked together in coupling.
Several researchers have found that the RAPD test is an effective technique for locating markers connected to agronomically significant traits that are introgressed during the development of close isogenic lines.
In tomato (Lycopersicon sp.), lettuce (Lactuca sp.), and common bean (Phaseolus vulgaris), disease resistance genes have been successfully linked to markers using RAPD analysis of NILs (non-isogenic lines).
High-density genetic mapping for numerous species of plants was developed swiftly through the effectiveness and speed of RAPD analysis.
Restriction Fragment Length Polymorphism – RFLP; Explained
Disadvantages:
The method’s main flaw is that as the profiling depends on the reaction conditions, it can differ between multiple laboratories. In addition, as each primer amplifies different discrete loci in the genome, profiles are unable to distinguish heterozygous from homozygous individuals.
Applications:
Although RAPDs and their associated modified markers have been used extensively in variability analysis and individual-specific genotyping, their use is less widespread due to issues like poor reproducibility, faint or fuzzy products, and difficulty scoring bands, which can result in wrong inferences.
Studies at the individual level (such as genetic identity) to studies involving closely related species and determination of genetic diversity have all been accomplished using RAPDs. To fill up the gaps left by other markers in gene mapping research, RAPDs have also been used.
Image reference: Cultivar identification and traceability, a molecular approach.
Published: 24-August-2023
Updated: 25-August-2023
Author: Ajmal Aseem
Graduated from the University of Kerala with B.Sc. Botany and Biotechnology. Attained Post-Graduation in Biotechnology from the Kerala University of Fisheries and Ocean Science (KUFOS) with the third rank. Conducted various seminars and attended major Science conferences. Done 6 months of internship in ICMR – National Institute of Nutrition, Hyderabad. 5 years of tutoring experience.