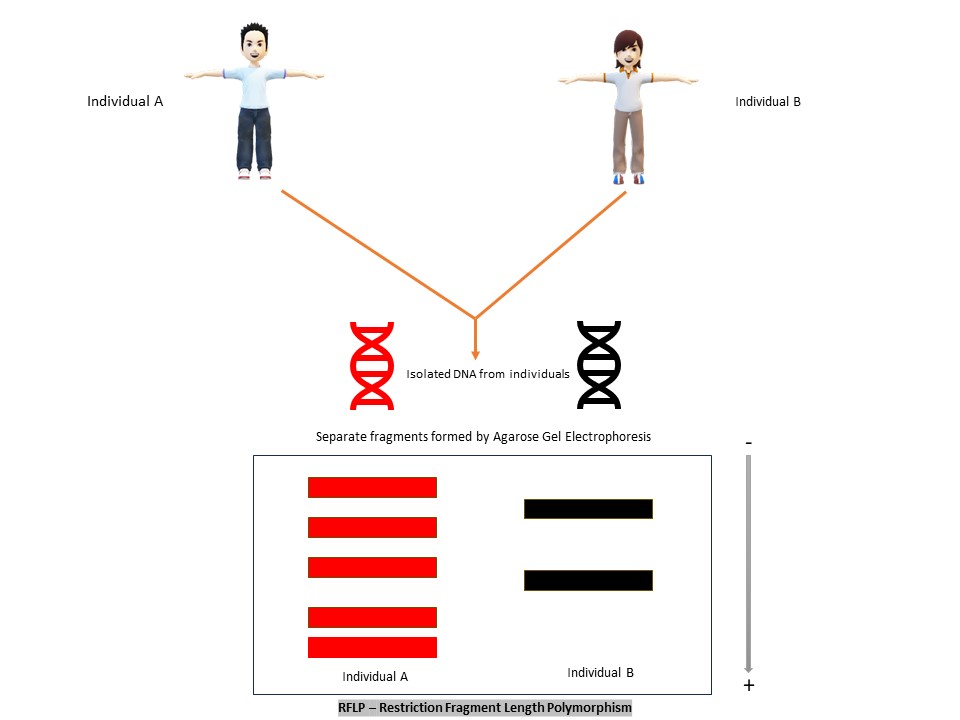
Restriction fragment length polymorphism markers were the first DNA-based genetic markers developed. The size of eukaryotic genomes hindered a simple identification of genetic variations in specific genes or sequences. The capability of complementary base pairing to reveal polymorphisms in sequences homologous to the probe enabled the development of methods to detect polymorphisms in these sequences. The approach provides a genetic system termed restriction fragment length polymorphism. A discrepancy between two or more samples of homologous DNA molecules that results from different restriction site locations is referred to as RFLP. When the DNA is digested by a restriction enzyme, the length of the fragments produced will vary depending on whether the two species have different lengths between the sites of cleavage of certain restriction endonucleases. It is possible to distinguish between species (and even strains) by the differences in the produced patterns.
Southern blotting, named after its creator E. M. Southern (1975), is a technique used to transfer and chemically bind fractionated DNA to a nylon membrane to solve this issue.
In RFLP analysis, the DNA sample is broken down into fragments by restriction enzymes and separated based on their lengths on an agarose gel electrophoresis. The DNA in such a gel can be observed by staining it with ethidium bromide, although distinct segments are often obscured by the abundance of restriction fragments of all sizes. Southern blotting, named after its creator E. M. Southern (1975), is a technique used to transfer and chemically bind fractionated DNA to a nylon membrane to solve this issue.
In-Depth
Combining the DNA fragments bound to the nylon membrane with a radioactively or fluorescently labelled DNA probe can detect specific DNA fragments. When individuals are tested, variable sizes or lengths of restriction fragments are frequently produced. It is possible to identify plant species, genotypes, and in some cases, individual plants using this sort of polymorphism. A radioactive isotope or alternate non-radioactive stains, like digoxigenin or fluorescein, may be used to label the probe. Most of these probes are single locus species-specific probes that range in size from 0.5 to 3.0 kb and were derived from a cDNA library or a genomic library. Several investigations show the opposite, despite the possibility that probes from cDNA libraries are more variable than those from genomic libraries
Advantages
The RFLP markers exhibit codominant inheritance, relatively high polymorphism, and great reproducibility. The RFLP markers are considered to be superior due to their widespread distribution throughout the eukaryotic genome and their high heritability and locus specificity. The approach also offers the chance to screen numerous samples simultaneously. DNA blots can be repeatedly examined by being stripped and reprobed (typically 8–10 times) with various RFLP probes.
Disadvantages
The method is not particularly popular because it is time-consuming, expensive, and just one of many markers that may be polymorphic, which is extremely inconvenient, especially for crosses between closely related species. Due to the huge amounts of pure, high molecular weight genomic DNA needed for DNA digestion and Southern blotting, the usefulness of RFLPs has been limited. The analysis is relatively risky and expensive as radioactive isotopes are used. The process is made more complex by the requirement for prior sequence knowledge for probe production. Their use in identifying point mutations within the locations where they detect polymorphism is constrained by their inability to detect single base changes.
Applications
Restriction Fragment length polymorphism is a powerful tool for the identification of organisms to the level of species, strains, varieties, or individuals. In linkage studies, RFLPs are frequently employed to produce maps for quantitative trait locus (QTL) research and to identify minor variations among populations in population genetics. They have also been employed as fingerprinting tools for investigations on diversity and hybridization, as well as to look into relationships between closely related taxa or at the intraspecific level. They have been applied to DNA testing for genetic abnormalities, paternity checks, and criminal investigations as well as to identify diseases.
Graduated from the University of Kerala with B.Sc. Botany and Biotechnology. Attained Post-Graduation in Biotechnology from the Kerala University of Fisheries and Ocean Science (KUFOS) with the third rank. Conducted various seminars and attended major Science conferences. Done 6 months of internship in ICMR – National Institute of Nutrition, Hyderabad. 5 years of tutoring experience.