Sanger sequencing, the primary approach of DNA sequencing since its inception in the 1970s, was used to complete the sequencing of the human genome in 2003. The need for rapid, cost-effective, and reliable analysis of vast amounts of genomic DNA is driving a massive increase in the demand for sequencing. With the use of contemporary sequencing methods known as Next Generation Sequencing, the entire human genome may now be sequenced in just a few hours. In this article, the concepts underpinning next-generation sequencing will be assessed, as well as its impacts on numerous scientific fields.
Sanger sequencing and Next Generation Sequencing (NGS) have a similar underlying principle. The genomic strand is fragmented, and the bases in each fragment are identified by emitted signals when the fragments are ligated against a template strand. The Sanger method has limitations in throughput, scalability, and resolution since it required various steps for sequencing, separation (by electrophoresis), and detection, making it challenging to automate sample preparation. The NGS methodology employs array-based sequencing, which combines the Sanger sequencing techniques to execute millions of reactions concurrently, resulting in extremely high speed and throughput at less expense.
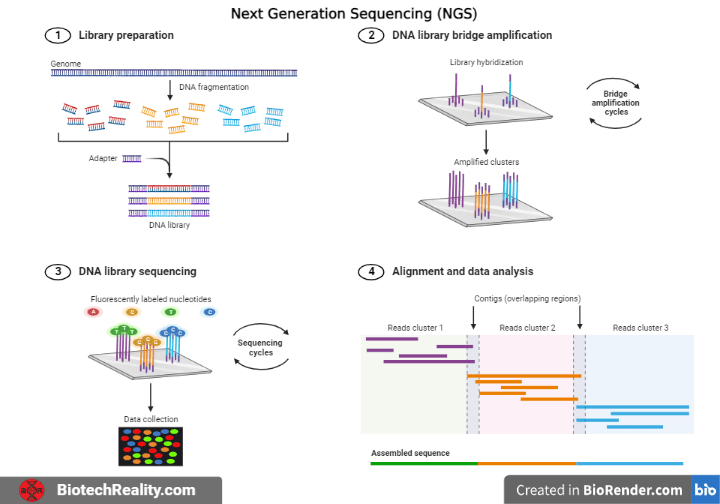
The following three processes are common to all next-generation DNA sequencing techniques:
1. Library Preparation: DNA is randomly fragmented to generate libraries, which are then ligated together using specific linkers.
2. Amplification: PCR and clonal amplification techniques are used to amplify the library.
3. Sequencing: DNA is sequenced using one of several methods.
4. Data Analysis: Bioinformatics pipelines are used to process the generated data in order to align the reads, find deviations, and assemble the entire genome.
Library preparation
Firstly, DNA is fragmented either enzymatically or by sonication to create smaller strands. Short and double stranded pieces of Synthetic DNA called Adaptors are then ligated to these fragments using DNA ligase enzyme. The adaptors enable the sequence to become bound to a complementary counterpart.
In order to attach the blunt end to the blunt terminated DNA, adaptors are synthesized with one end that is “sticky” and the other end that is “blunt”. Base pairing between molecules may become a concern as a result, which might culminate in dimer formation. The chemical composition of DNA is utilized to prevent this since ligation occurs between the 3′-OH and 5′-P ends. By removing the phosphate from the sticky end of the adaptor and therefore creating a 5′-OH end instead, the DNA ligase is unable to form a bridge between the two termini.
Amplification
It is necessary to use library amplification to make the received signal from the sequencer sufficiently accurate to be reliably detected. Enzymatic amplification is susceptible to processes like “biasing” and “duplication,” which can result in the preferential amplification of particular library fragments. Instead, a number of amplification methods use PCR to produce many DNA clusters in enormous quantities.
Sequencing
Various companies have developed multiple types of Next Generation Sequencing competitive techniques (Pyrosequencing, Sequencing by ligation (SOLiD), Reversible terminator sequencing (Illumina))
Applications of NGS:
Researchers have been able to gather enormous amounts of genomic sequencing data through next-generation sequencing. This technology has a wide range of applications including the diagnosis and understanding of complex diseases, whole-genome sequencing, epigenetic analysis, mitochondrial sequencing, transcriptome sequencing, understanding how altered expression of genetic variants affects an organism, and exome sequencing – mutations in the exome are thought to contain up to 90% of mutations in the human genome, which results in disease.
Gene therapy in cancer treatment focuses on methods like antisense RNA, which prevents the synthesis of targeted proteins, and suicide gene therapy, which introduces genes to selectively kill cancer cells. The challenge lies in delivering these genes precisely to avoid harming healthy cells. Sequencing tumor genomes enables tailored chemotherapy and personalized medicine, revolutionizing diagnostics and treatment planning.
The decreasing cost of DNA sequencing leads to its wider adoption, but it also presents challenges. Processing and storing the vast amounts of sequencing data pose computational hurdles. Ethical concerns arise regarding ownership and security of individuals’ DNA data, as it can be misused by insurance companies, mortgage brokers, or employers.
Additionally, while sequencing can identify disease risks, issues remain regarding patient awareness and the availability of effective treatments.
Title Image Source: Created in BioRender.com
Graduated from the University of Kerala with B.Sc. Botany and Biotechnology. Attained Post-Graduation in Biotechnology from the Kerala University of Fisheries and Ocean Science (KUFOS) with the third rank. Conducted various seminars and attended major Science conferences. Done 6 months of internship in ICMR – National Institute of Nutrition, Hyderabad. 5 years of tutoring experience.
Read More






One thought on “Next Generation Sequencing (NGS) and its application in genomics”
Comments are closed.