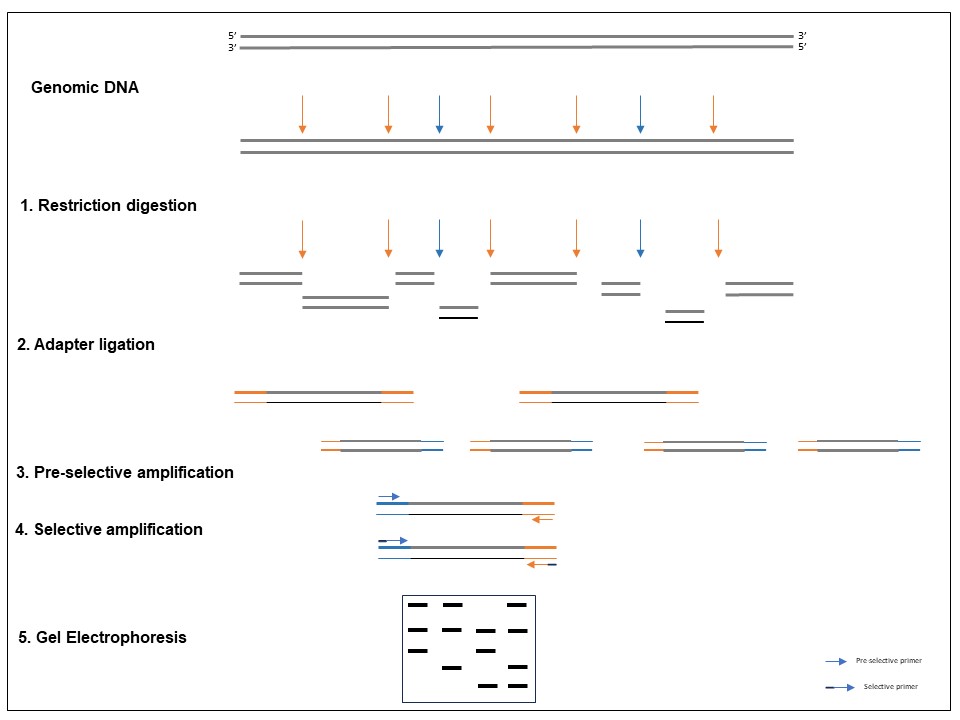
Amplified fragment length polymorphism PCR is a PCR-based tool used in genetics research, DNA fingerprinting and in the practice of genetic engineering. In short, AFLP is an intermediate stage between RFLP and PCR.
A complex combination of DNA fragments acquired after the digestion of genomic DNA with restriction endonucleases is selectively amplified by AFLP. By using polyacrylamide gel electrophoresis (PAGE) or capillary electrophoresis, polymorphisms can be identified from variations in the length of the amplified fragments.
In AFLP, genomic DNA is first cut with restriction enzymes, and then adaptors are joined to the sticky ends of the restriction fragments. Then, a subset of the restriction fragments is amplified using primers complementary to the adapter and some of the restriction site fragments.
The amplified fragments can be observed using autoradiography or fluorescence techniques on denaturing polyacrylamide gels. AFLPs are DNA fragments (80–500 bp) produced by restriction enzyme digestion, oligonucleotide adapter ligation to the digestion products, and targeted PCR amplification.
RFLP and PCR are therefore both used in AFLPs. Variations in the restriction sites or the surrounding area lead to changes in the AFLP banding profiles.
The AFLP method produces fragments from numerous genomic locations concurrently (about 50–100 fragments per reaction), sorted by polyacrylamide gel electrophoresis and often graded as dominant markers.
Also read: Restriction Fragment Length Polymorphism – RFLP; Explained
Advantages
AFLP provides a lot of advantages, including the ability to amplify between 50 and 100 fragments at once and be produced in larger quantities. It also has improved repeatability, resolution, and sensitivity at the whole genome level.
Additionally, amplification does not require knowledge of the sequence in the past. As a result, AFLP has proven to be very helpful in the study of taxa such as bacteria, fungi, and plants, where there is still much to learn about the genetic makeup of distinct organisms.
Automatic sequencers can analyze AFLPs, although certain systems encounter software issues that affect the score of AFLPs. Due to its ability to reveal a large number of polymorphic markers in a single reaction, AFLP has become the primary tool in genetic marker technology.
Disadvantages
This method has the drawbacks of being labour-intensive, having medium repeatability, having difficulty in identifying alleles, and having high operating and developing expenditures. Purified, high molecular weight DNA, allele dominance, and the potential non-homology of co-migrating fragments from various loci are requirements for AFLP.
Applications
The majority of AFLP fragments map to specific locations on the genome, can concurrently identify multiple polymorphisms in several genomic areas, and can therefore be used as markers in both genetic and physical mapping. The method can be applied to gene mapping as well as to the subspecies level to discern closely related individuals.
Applications for AFLP in plant mapping involve developing linkage groups in crosses, saturating locations with markers for gene landing operations, and figuring out how closely related or variable different cultivars are.
For genetic investigations, molecular markers are more reliable than morphological characteristics since they are unaffected by environmental factors. Due to the prevalence of null alleles, AFLP seems to be more applicable to intraspecific than to interspecific investigations.
In genetic investigations such as evaluating biodiversity, analyzing germplasm collections, genotyping individuals, and analyzing genetic distance, AFLP markers are useful.
For specific purposes (such as polymorphism screening, QTL analysis, and genetic mapping), the direct manipulation of AFLP fragment development can be performed considering the wide variety of restriction enzymes and matching primer combinations that are readily available.
Graduated from the University of Kerala with B.Sc. Botany and Biotechnology. Attained Post-Graduation in Biotechnology from the Kerala University of Fisheries and Ocean Science (KUFOS) with the third rank. Conducted various seminars and attended major Science conferences. Done 6 months of internship in ICMR – National Institute of Nutrition, Hyderabad. 5 years of tutoring experience.